This repository contains a reciprocal BLAST program for filtering down BLAST results to best bidirectional hits. It also contains a toolkit for finding and visualizing BLAST hits for gene clusters within multiple bacterial genomes.
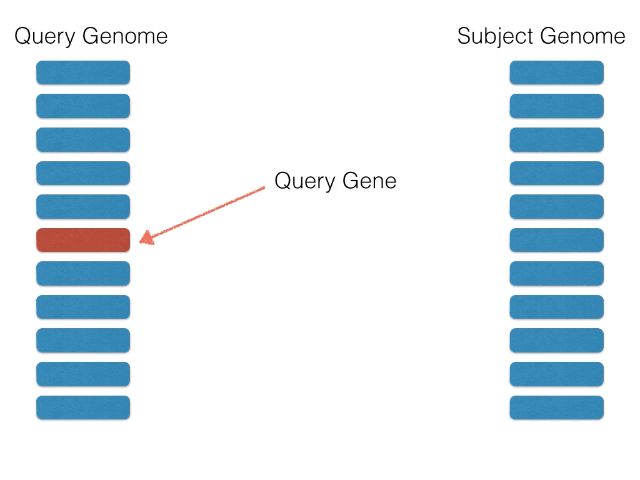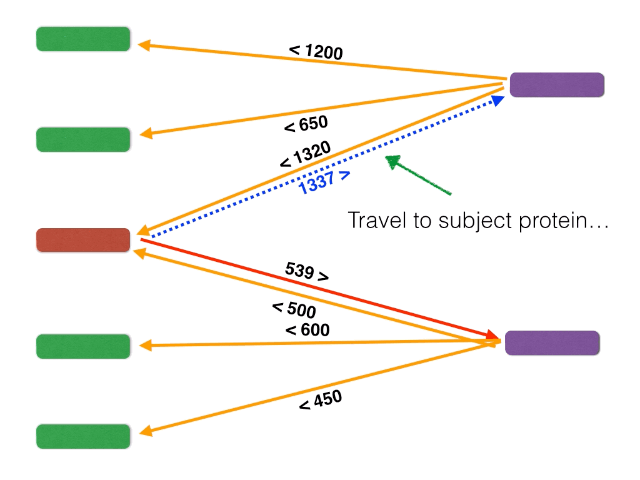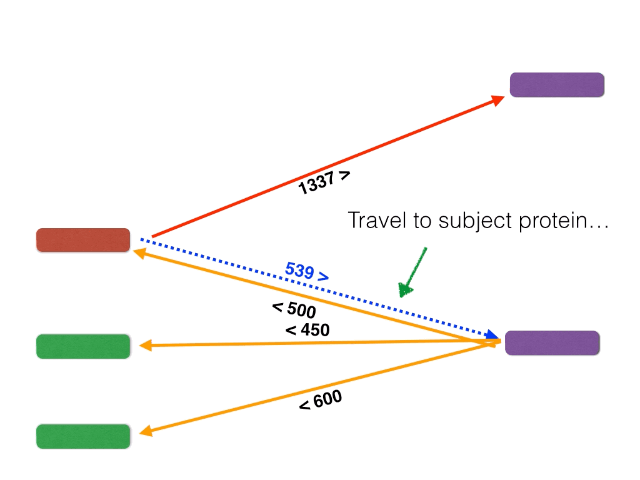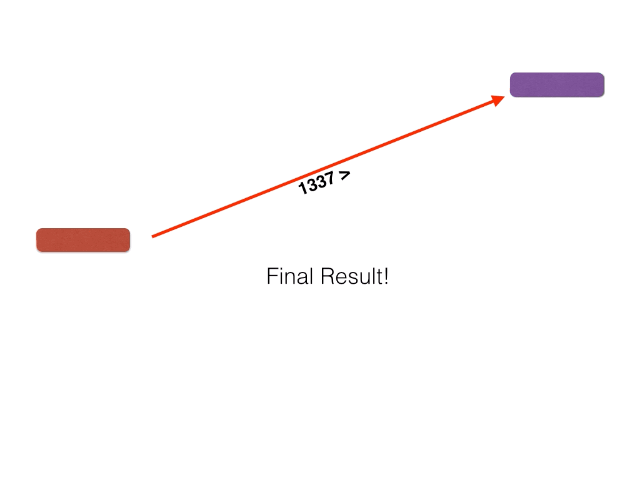
- BackBLAST.py - A script that uses NCBI BLAST to search for gene clusters within a within a bacterial genome genome. Non-orthalagous genes are filtred out by identifying and extracting only Bidirectional BLAST Hits using a graph-based algorithm. The algorithm is illustrated below:
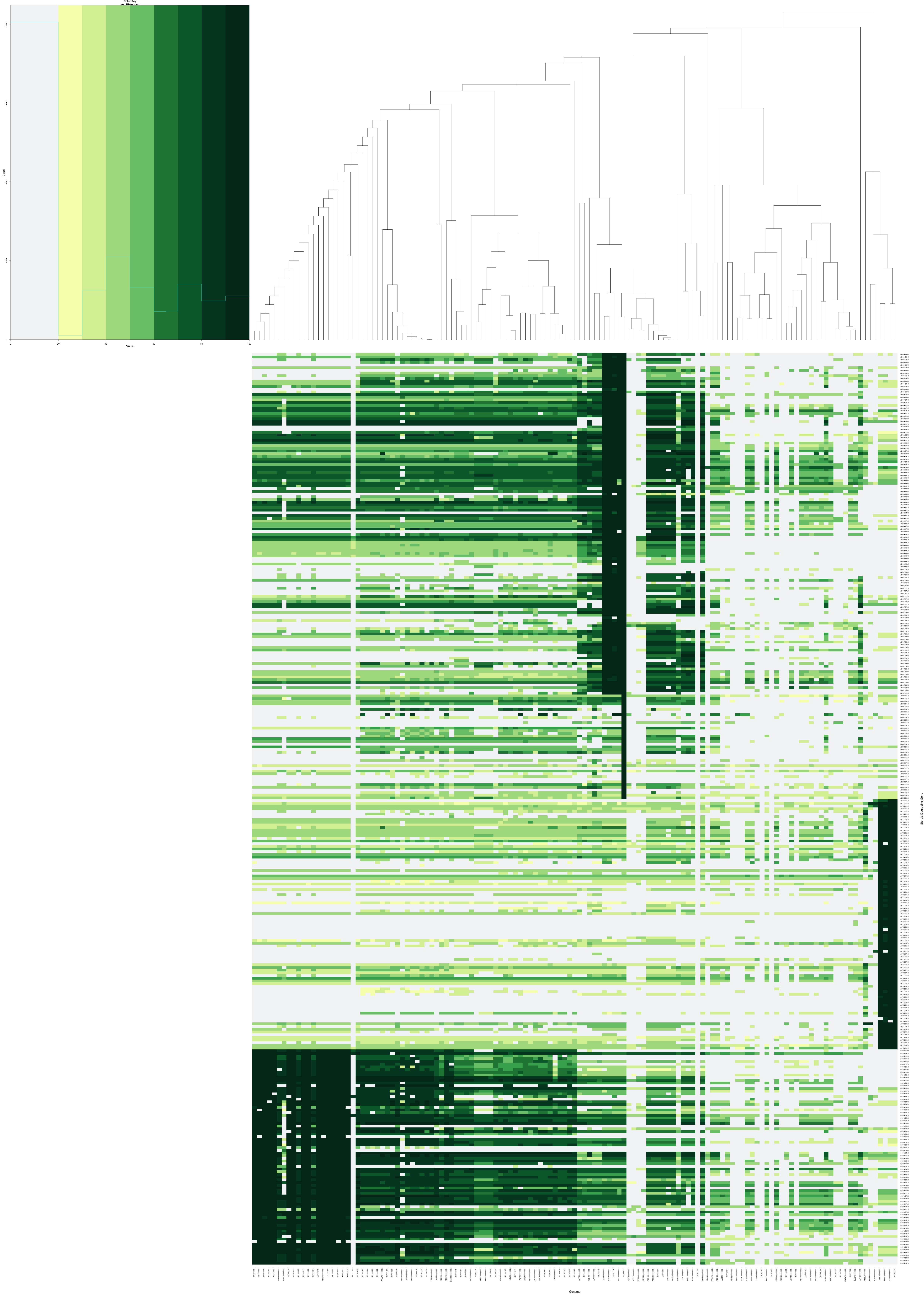
- Visualization - This repository also includes tools for visualizing the results from BackBLAST.py in the form of a R heatmap. Here is an example:
For more thorough descriptions and information on usage please check the [wiki!] (https://github.com/LeeBergstrand/BackBLAST-Gene-Cluster-Finder/wiki)