A Parallel Unmixing-Based Content Retrieval System for Distributed Hyperspectral Imagery Repository on Cloud Computing Platforms
As the volume of remotely sensed data grows significantly, content-based image retrieval (CBIR) becomes increasingly important, especially for cloud computing platforms that facilitate processing and storing big data in a parallel and distributed way. We proposes a novel parallel CBIR system for hyperspectral image (HSI) repository on cloud computing platforms under the guide of unmixed spectral information, i.e., endmembers and their associated fractional abundances, to retrieve hyperspectral scenes. However, existing unmixing methods would suffer extremely high computational burden when extracting meta-data from large-scale HSI data. To address this limitation, we implement a distributed and parallel unmixing method that operates on cloud computing platforms in parallel for accelerating the unmixing processing flow. In addition, we implement a global standard distributed HSI repository equipped with a large spectral library in a software-as-a-service mode, providing users with HSI storage, management, and retrieval services through web interfaces. Furthermore, the parallel implementation of unmixing processing is incorporated into the CBIR system to establish the parallel unmixing-based content retrieval system.
clone the repository:
$ git clone [email protected]:ZpWaitingForSunshine/CBIR.git
If this is your first time using Github, review http://help.github.com to learn the basics.
You can also download the zip file containing the code from https://github.com/ZpWaitingForSunshine/CBIR/archive/master.zip
This project includes three parts: the frontend code, the backend code, and the parallel algorithm code.
This application is depolyed on the cloud platform. First, you should have an Openstack platform. Then, create several virtual machines including a Spark cluster, a HDFS cluster, a application server(as a file server too), and a MySQL server.
In Detail, the software required as shown in the following list:
| Software | Release Version |
|---|---|
| Spark | 2.3.3 |
| Hadoop | 2.7.3 |
| OpenStack | Queens |
| MySQL | 5.7.1 |
| Java | 1.8.0_201 |
| Scala | 2.11.8 |
| Tomcat | 9.0 |
| Spring | 4.3.1.RELEASE |
| MyBatis | 1.3.1 |
- For the frontend:
The frontend is based on a production-ready frontend solution named vue-element-admin. You can follow the guide to run the frontend project.
Simply run with:
# enter the project directory
$ cd hsicloud_frontend
# install dependency
$ npm install
# develop
$ npm run dev
- For the backend:
The backend was developed based on Spring, SpringMVC, MyBatis (SSM), and MySQL.
First, import the data file hsicloud.sql to the MySQL server.
Secondly, upload the unmixing jar named PPI_SAD_SCLS.jar to the HDFS.
Finally, import the code in IDEA as maven project. After adjusting JRE system library and dependencies, run the application on Tomcat.
- For the parallel algorithm code
Open the project in IDEA, and import necessary libraries such jars in Spark. Package the application as a jar, and submit Spark jobs to the Spark cluster
Note: set the IP until adaptive
In what follows, we provide a simple step-by-step example to demonstrate how to perform a simple hyperspectral image retrieval in our system.
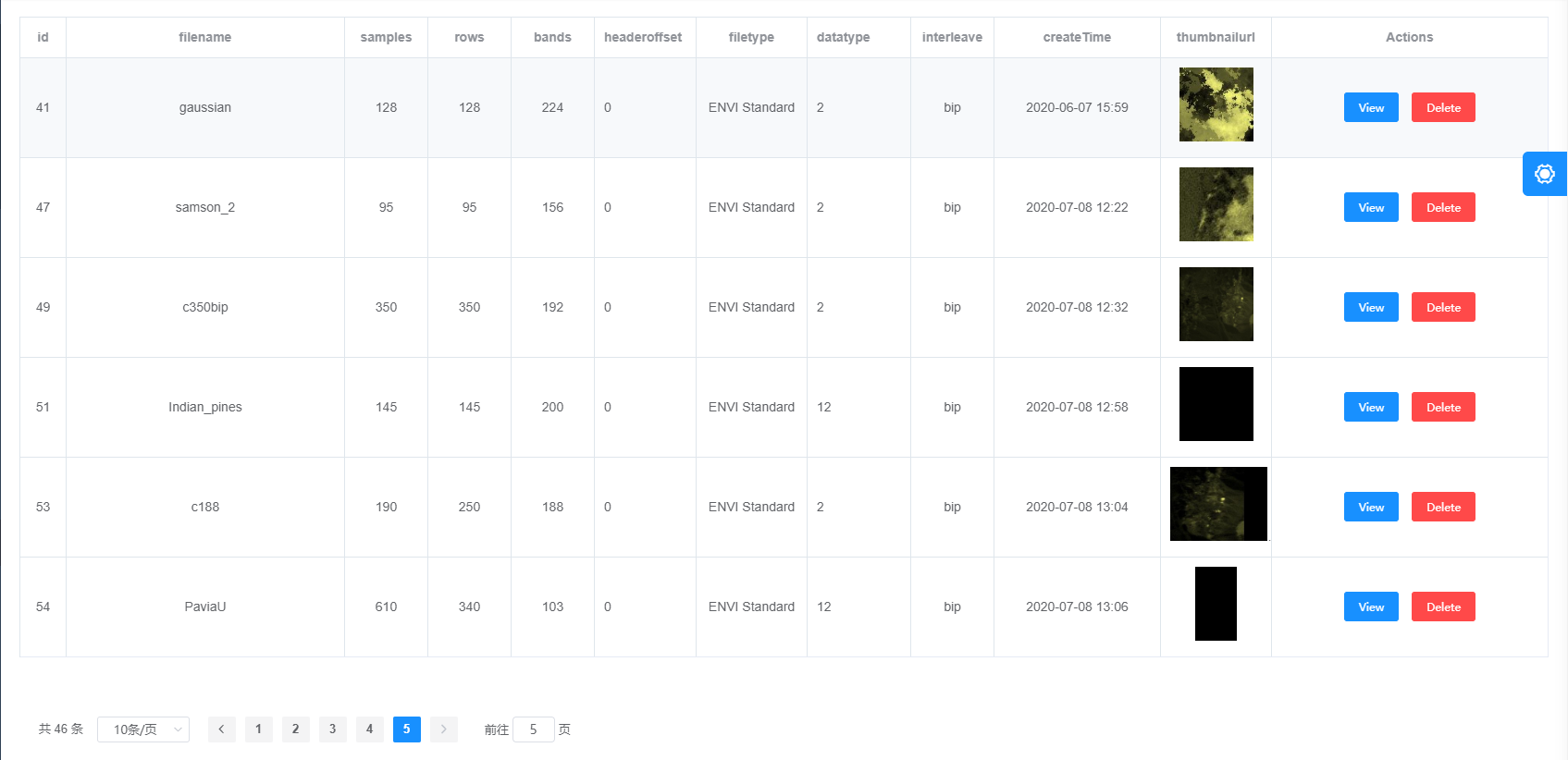
Figure 1
Figure 1 shows the list of HSIs in our system with their general metadata.
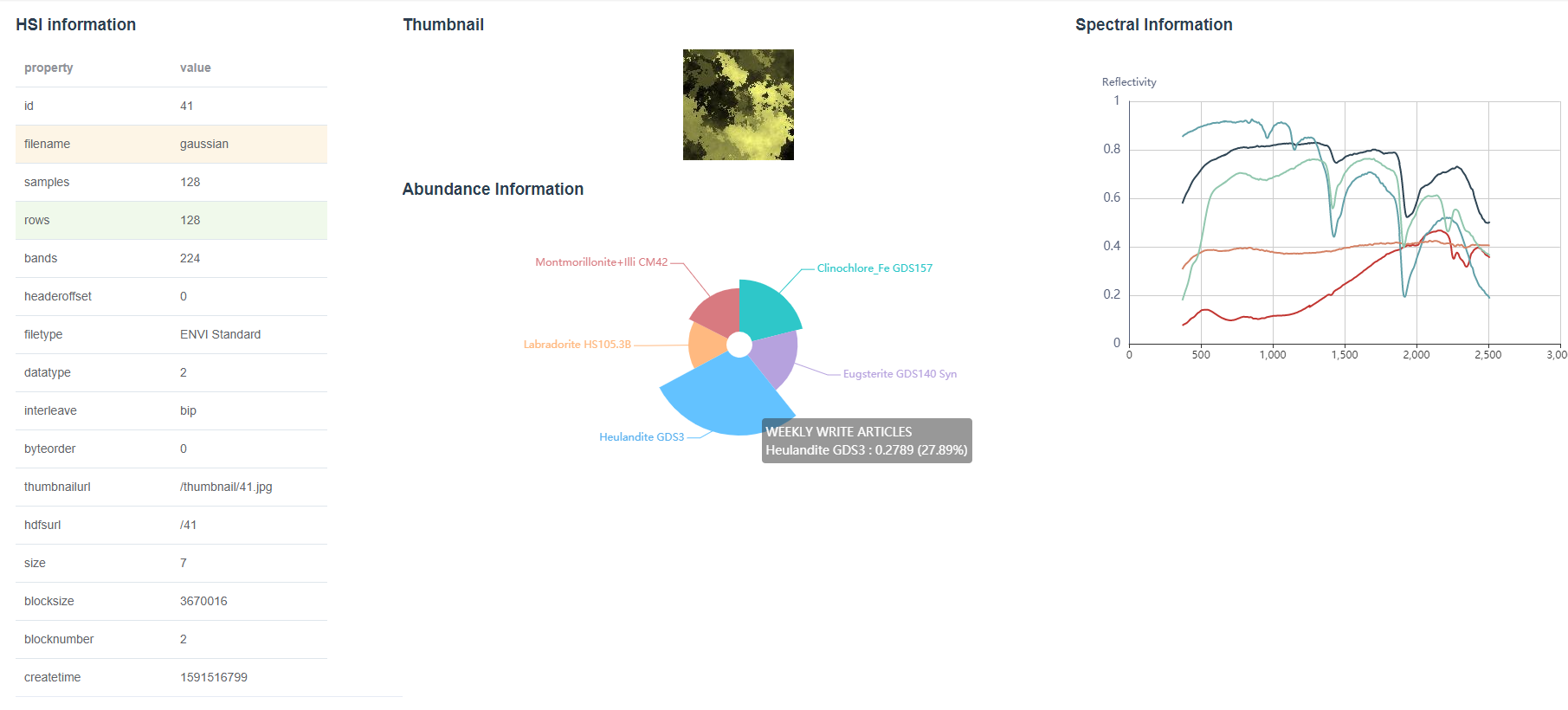
Click the View command button and then the HSI’s information including all metadata, thumbnail, its endmembers’ spectrum, and the ratio of every endmember will be displayed in detail as shown in Figure 2.
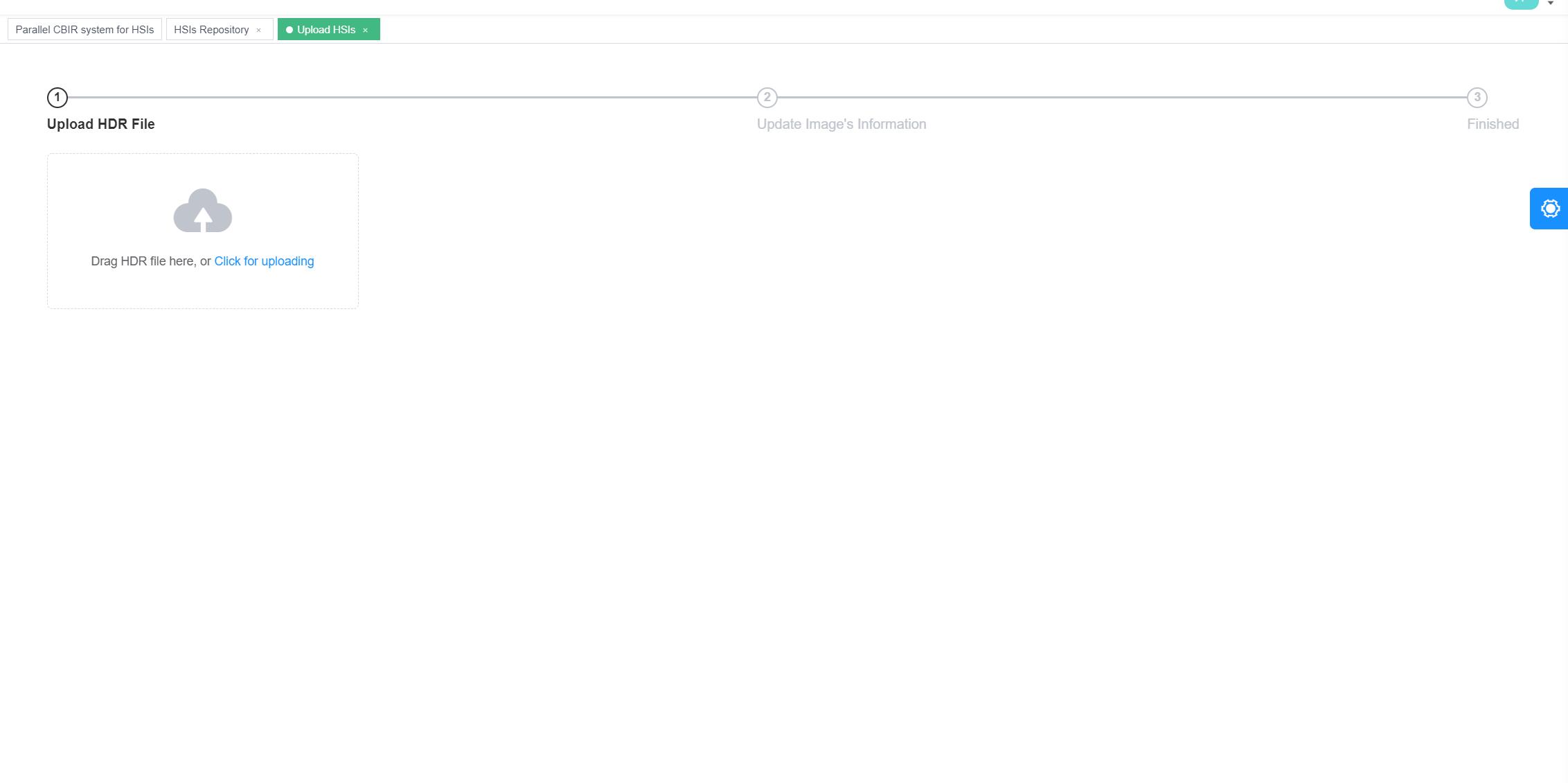
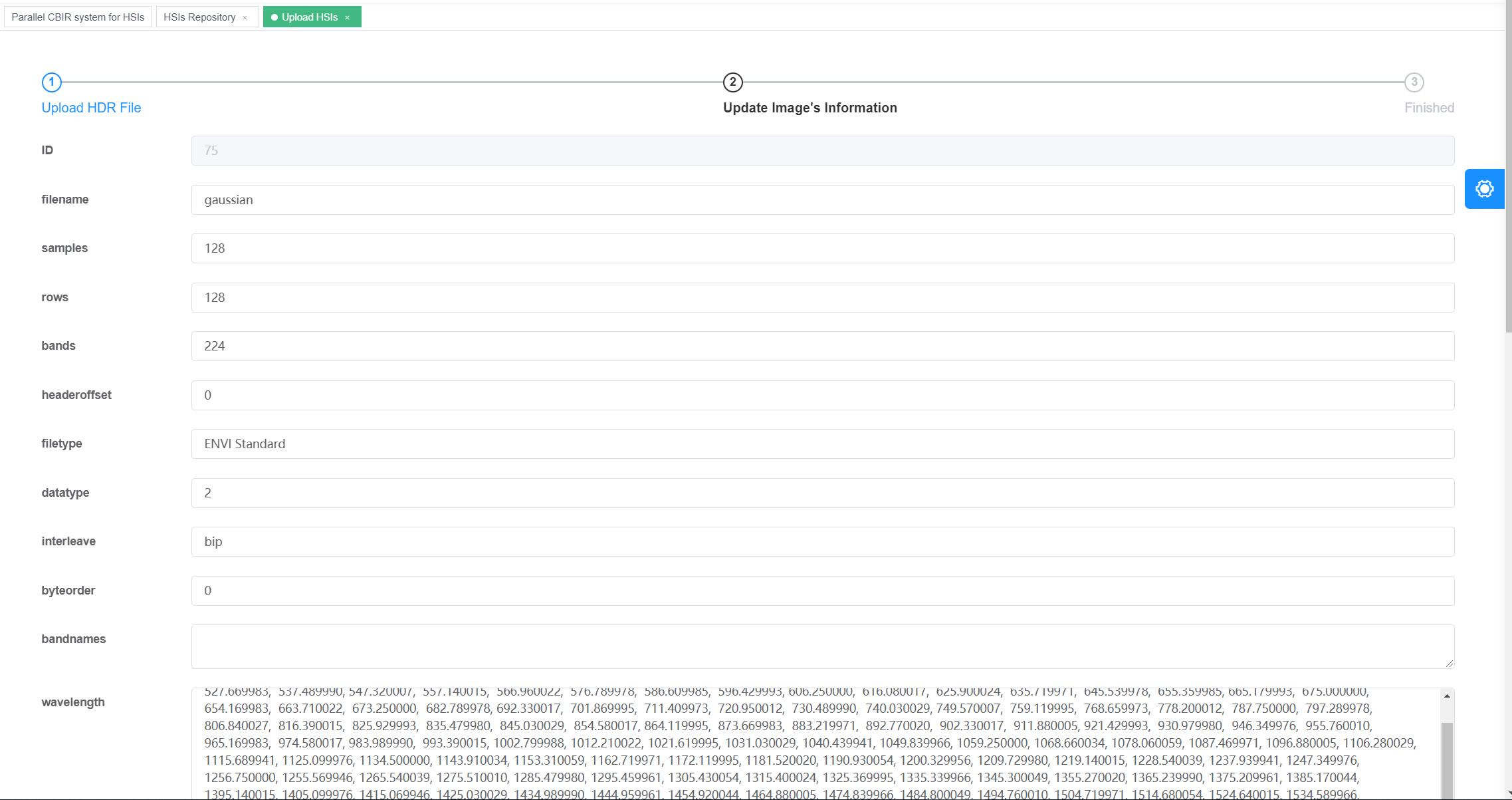
To expand the HSI repository, the system provides an interface for uploading as shown in Figure 3 and 4. Once an HSI is uploaded, the users can decide which unmixing algorithms to extract the spectral information.
As shown in Figure 5, the PPI, SAD and SCLS algorithms based on Spark were chose which can be executed in appointed cluster in the cloud framework. In addition, the parameters including Driver-Memory, Executor-Memory, and Executor-Cores for task execution can also be specified in the web page.
After the unmixing algorithms have been executed, the HSI spectral information obtained as a complement will be automatically cataloged in the database table abundance.
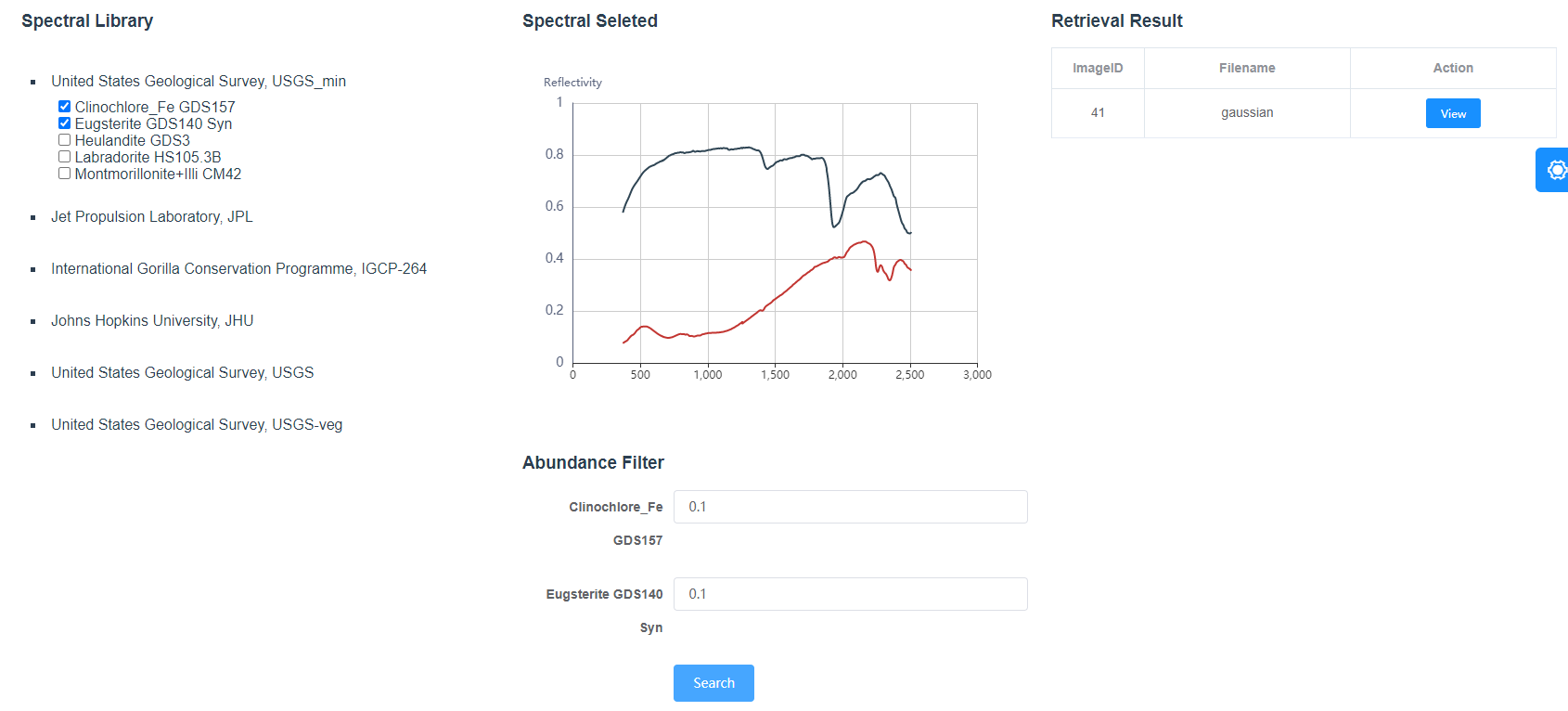
Figure 6 shows a querying example, in which we specified two spectral signatures (clinochiore_Fe GDS157 and Eugsterite GDS140 Syn) from United States Geological Survey (USGS) library as the query criteria. The minimum abundance is set to 0.1 that means the HSIs we are looking forward to contain at least 10% clinochiore_Fe GDS157 and10% Eugsterite GDS140 Syn. The query result will be shown as a list and corresponding HSI’s information can be viewed by click the View button. In this case, the HSI whose id is 41 contains 21.05% clinochiore_Fe GDS157 and 18.25% Eugsterite GDS140 Syn which satisfies the query criteria accurately.
If you have questions or comments need to reply, please contact with us via email:[email protected]